nf-core/genomeqc
Compare the quality of multiple genomes, along with their annotations.
Introduction
This document describes the output produced by the nf-core/genomeqc.
The directories listed below will be created in the results directory after the pipeline has finished. All paths are relative to the top-level results directory.
Pipeline overview
The pipeline is built using Nextflow and processes data using the following steps:
- NCBI genome download - Download genomes and their annotations from RefSeq
- Genome quality metrics:
- Annotation quality metrics:
- AGAT sp_statistics - Gene statistics
- AGAT sp_keep_longest_isoform - Filter longest isoform from GXF file
- Gene overlaps - Find overlapping genes (sense and antisense)
- Decontamination:
- FCS-GX - Foreign genome contamination screening
- FCS-adaptor - Adaptor and vector contamination screening
- FCS-adaptor clean genome Removal of contamination from assembly
- Tiara - Sequence classification (domain and organelle level)
- GffRead - Extract longest isoform from FASTA file
- BUSCO - Genome completeness based on single copy markers
- Orthofinder - Phylogenetic orthology inference
- Tree summary - Phylogenetic summary plot
- MultiQC - Aggregate report describing results and QC from the whole pipeline
- Pipeline information - Report metrics generated during the workflow execution
Decontamination
FCS-GX
FCS-GX is a module in NCBI’s FCS (Foreign Contamination Screening) toolkit designed to detect contaminant sequences of foreign organisms from genome assemblies.
It generates a report with assembly sequence classifications, contamination summaries, and cleaning recommendations.
Output files
decontamination/fcs-gx<assembly>_<taxid>.fsc_gx_report.txt: Summary report with sequence classification and cleaning recommendations.<assembly>_<taxid>.taxonomy.rpt: Detailed breakdown of sequence classification.
FCS-Adaptor
FCS-Adaptor is part of the NCBI’s FCS toolkit. It’s specifically designed to detect adaptor and vector contamination that sometimes remain in genome assemblies.
It generates a report with a list of sequences flagged as adaptor or vector matches, cleaning recommendations, and the cleaned genome assembly.
Output files
decontamination/fcs-adaptor<assembly>.cleaned_sequences.fa.gz: Genome assembly with contaminant regions removed.<assembly>.fsc_adaptor_report.txt: Summary report with flagged sequences and cleaning recommendations.
We recommend ignoring the cleaned genome assembly output by this module, as the vector and adapter removal is done by the FCS-GX clean genome module (see below).
FCS-GX clean genome
FCS-GX clean genome is based on a command of the NCBI’s FCS toolkit which applies the recommended cleaning actions to the genome assembly based on the screening results.
It outputs a cleaned version of the genome assembly based on the recommended actions from FCS-GX and FCS-Adaptor, with the contaminant sequences removed and sequences with local contaminants trimmed.
Output files
decontamination/cleaned_genome<assembly>.cleaned.fasta: Genome assembly with contaminant sequences removed and contaminant regions trimmed.<assembly>.contaminants.fasta: Sequences classified as contaminants.
Tiara
Tiara is a deep learning–based classifier designed to identify eukaryotic, archaeal, and bacterial sequences, as well as organelle genomes.
It outputs a report with each sequence of the genome assembly labelled as Eukarya, Archea, Bacteria, organelle or unknown.
Output files
decontamination/tiara<assembly>.txt: Report with sequence classifications.log_<assembly>.txt: Log file with classification statistics and model information.
NCBI genome download
NCBI genome download is a tool for downloading assemblies from the NCBI FTP site.
It inputs RefSeq IDs and downloads the respective assembly and annotation in FASTA and GFF formats. If local files are provided, this step is skipped.
Output files
ncbigenomedownload/<assembly>.fa.gz: Genome assembly in FASTA format.<assembly>.gff3.gz: Annotation in GFF format.
This directory will only be present if --save_assembly flag is set.
Quast
Quast provides different quality metrics about the genome assembly. It computes contiguity stats (N50, N90), genome size, GC% content and number of sequences.
It generates a report in different formats, as well as an HTML file with in integrated contig viewer.
Output files
quast/<species_name>/icarus.html: Contig viewer in HTML formatreport.html: Assembly stats in HTML formatreport.pdf: Assembly stats in tsv formatreport.tsv: Assembly QC as HTML report
tidk
tidk is a tool to identify and visualise telomeric repeats from asseblies.
It will use a known telomeric repeat as input string, and will find occurrences of these sequence in windows across the genome.
Output files
tidk/<species_name>.tsv: Report with the number of repeats found in different number of windows<species_name>.svg: Plot with the repeat distribution
To run nf-core/genomeqc with tidk, the flag --run_tidk must be provided.
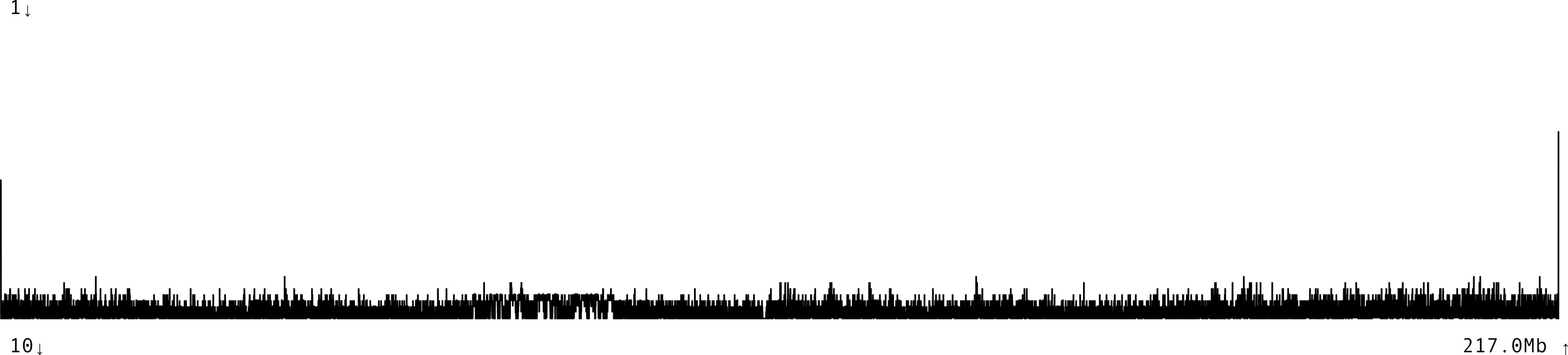
Merqury
Merqury uses k-mers from sequencing reads to evaluate the assembly quality and completness without the need of a high quality reference.
It generates a histogram relating k-mer counts in the read set to their associated counts in the assembly, as well as a completness report.
Output files
merqury/<assembly>.html: Contig viewer in HTML format<assembly>.html: Assembly stats in HTML format<assembly>.pdf: Assembly stats in tsv format<assembly>.tsv: Assembly QC as HTML report
To run nf-core/genomeqc with merqury, the flag --run_merqury must be provided.
AGAT sp_statistics
AGAT sp_statistics computes several annotation metrics such as number of genes, transcripts, exons, etc.
Output files
agat/<species_name>.stats.txt: Contig viewr in HTML format
AGAT sp_keep_longest_isoform
AGAT sp_keep_longest_isoform filters GXF file to keep the longest isoform per gene. Longest isoforms are recommended as input for both BUSCO and Orthofinder.
Output files
longest/<species_name>.longest.g*f: Contig viewr in HTML format
This directory will only be present if --save_longest_isoform flag is set.
Gene overlaps
Gene overlaps is a local module based on the R package GenomicRanges, used for manipulating genomic intervals. It finds the number of genes that are overlapping in the GXF file, which can be used as a metric to evaluate the quality of the annotation.
It outputs a brief report with information about the number of reads, the number of genes fully contained in sense direction and in the antisense direction, and the total number of overlapping genes.
Output files
longest/Count.<species_name>.tsv: Report in tsv format
GffRead
GffRead extracts the protein sequences using the genome assembly and annoation as input.
Output files
gffread/<species_name>.longest.fasta: Report in tsv format
This directory will only be present if --save_extracted_seqs flag is set.
BUSCO
BUSCO is a tool for assessing the quality of assemblies based on the presence of single copy orthotologues. It computes the compleness based on evolutionarily informed expectations of gene content, whether this single copy markers are present in single copy, duplicated, fragmented or absent.
It outputs a report with completness stats, a summarized table with these stats, and an ideaogram with single copy markers mapped against each chromosome or sequence.
Output files
busco/short_summary.specific.<busco_db>.<species_name>.fasta.txtCompletness report in tsv format<species_name>-<busco_db>-busco.batch_summary.txt: Summarized completness report in tsv format<species_name>_<lineage>.pngIdeogram with the location of single copy markers
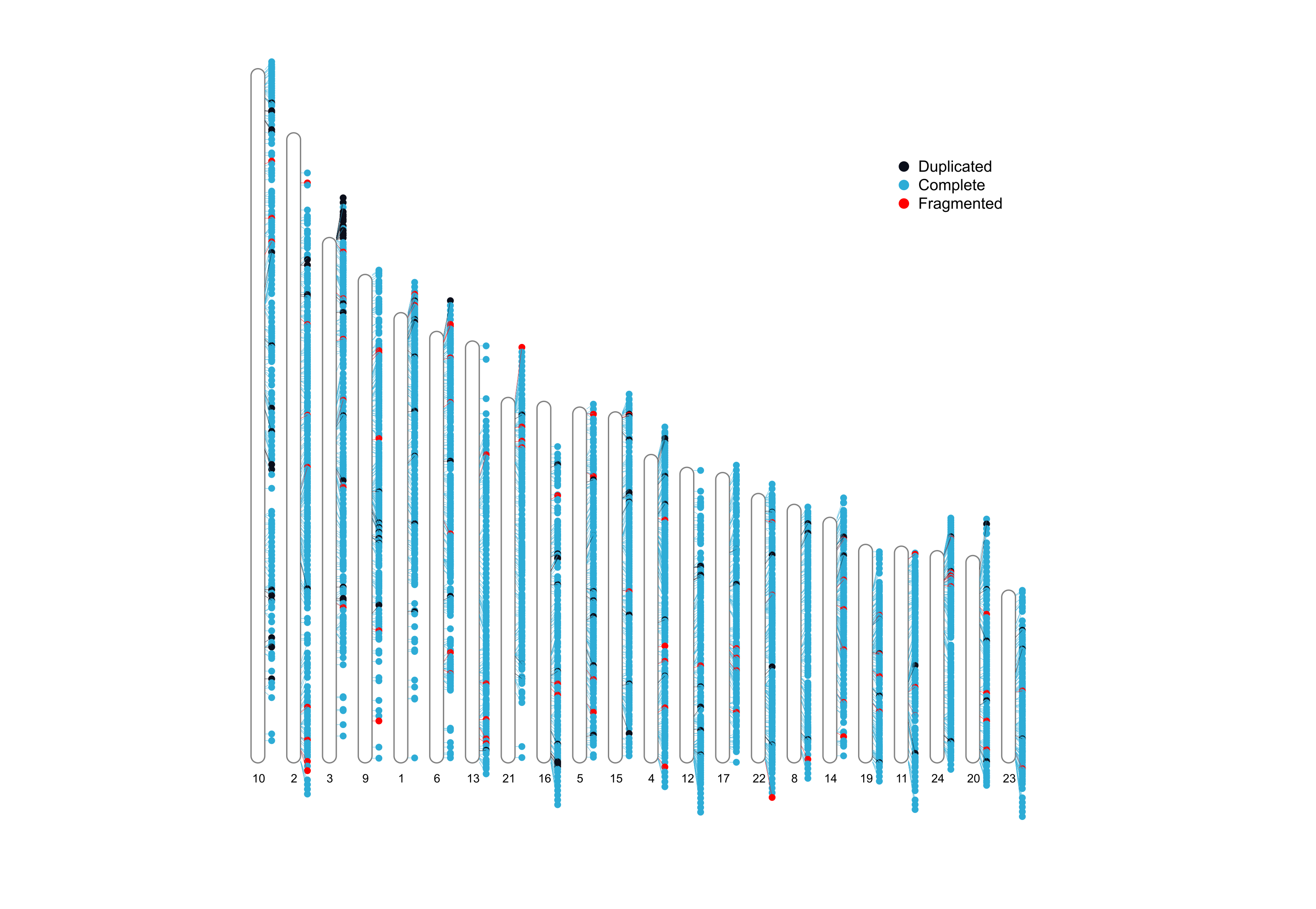
Orthofinder
Orthofinder finds groups of orthologous genes and uses these orthologous genes for phylogenetic inference.
It output a rooted species tree which is later used to present the quality stats of the assemblies.
Tree summary
Tree summary is a local module that takes the rooted trees species from Orthofinder, as well as the ouput statistics from the mentioned tools.
The idea of the tree summary is to give some phylogenetic context to the quality stats, which might help users when evaluating the integrity of the assemblies.
tree_summary/tree_plot.pdfTree summary with quality statistics
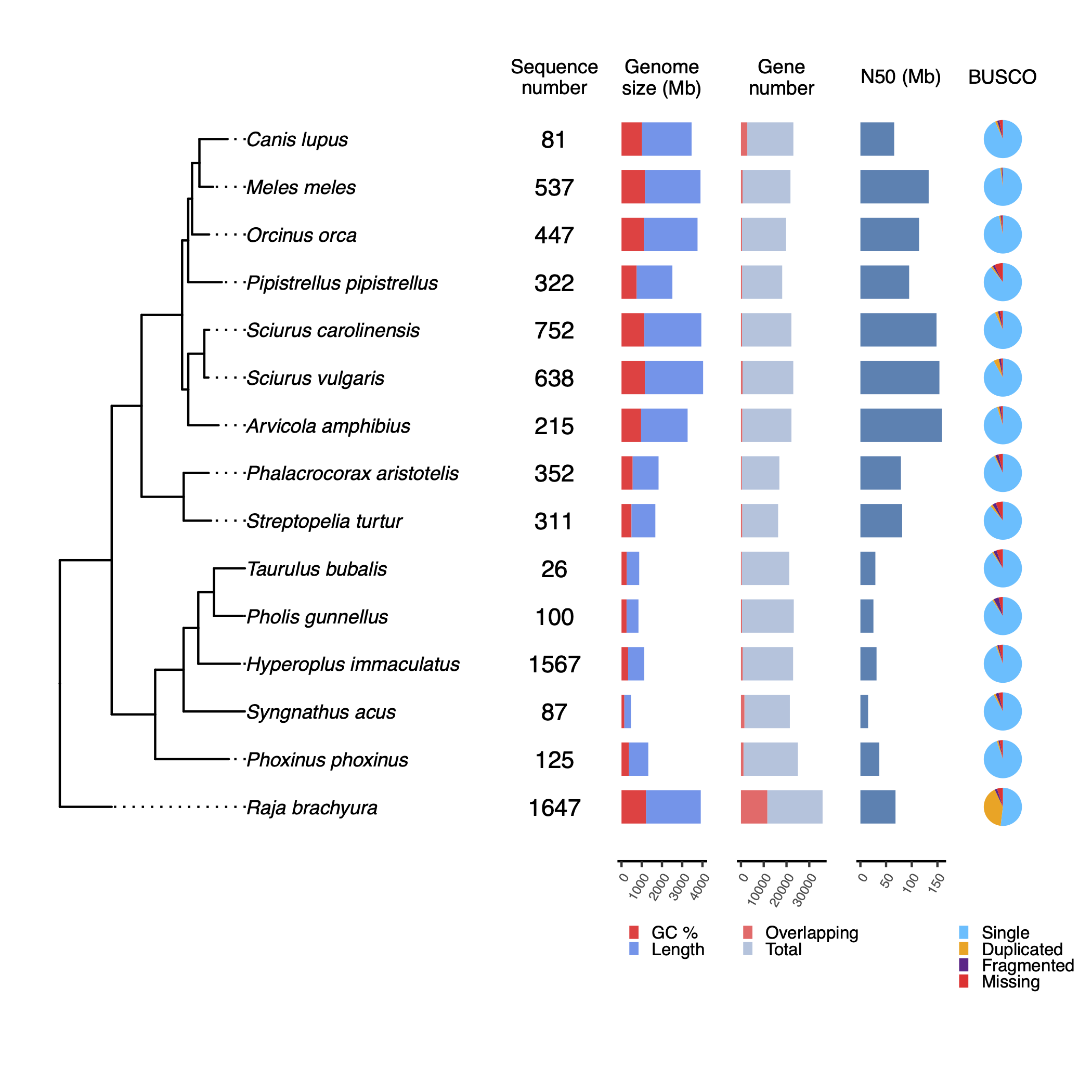
MultiQC
MultiQC is a visualization tool that generates a single HTML report summarising all samples in your project. Most of the pipeline QC results are visualised in the report and further statistics are available in the report data directory.
Results generated by MultiQC collate pipeline QC from supported tools e.g. FastQC. The pipeline has special steps which also allow the software versions to be reported in the MultiQC output for future traceability. For more information about how to use MultiQC reports, see http://multiqc.info.
Output files
multiqc/multiqc_report.html: a standalone HTML file that can be viewed in your web browser.multiqc_data/: directory containing parsed statistics from the different tools used in the pipeline.multiqc_plots/: directory containing static images from the report in various formats.
Pipeline information
Nextflow provides excellent functionality for generating various reports relevant to the running and execution of the pipeline. This will allow you to troubleshoot errors with the running of the pipeline, and also provide you with other information such as launch commands, run times and resource usage.
Output files
pipeline_info/- Reports generated by Nextflow:
execution_report.html,execution_timeline.html,execution_trace.txtandpipeline_dag.dot/pipeline_dag.svg. - Reports generated by the pipeline:
pipeline_report.html,pipeline_report.txtandsoftware_versions.yml. Thepipeline_report*files will only be present if the--email/--email_on_failparameter’s are used when running the pipeline. - Reformatted samplesheet files used as input to the pipeline:
samplesheet.valid.csv. - Parameters used by the pipeline run:
params.json.
- Reports generated by Nextflow: